Graph
Background
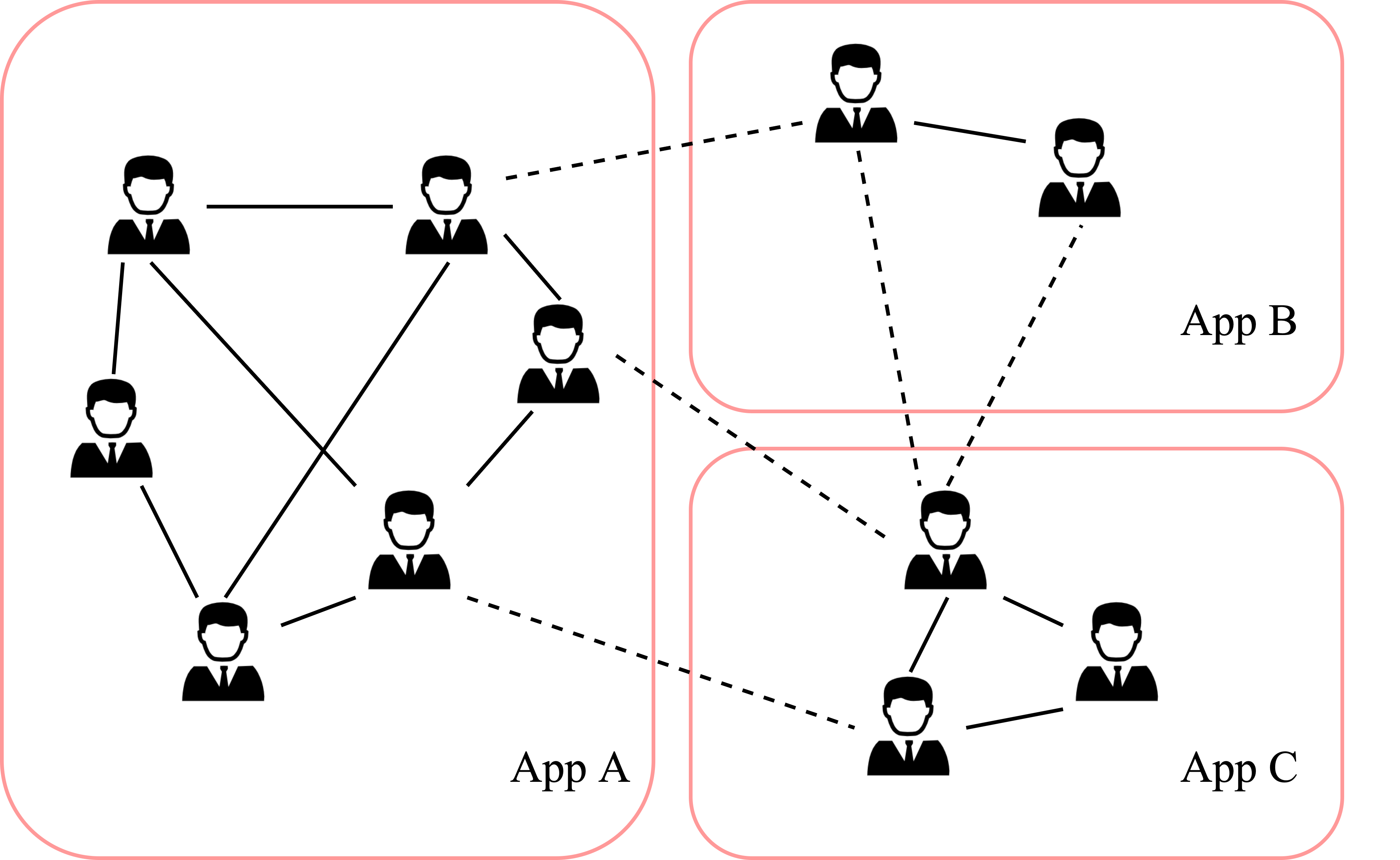
For privacy reasons, there are many graphs in scenarios that are split into different subgraphs in different clients, which leads to missing of the cross-client edges and data non.i.i.d., etc.
Not only in areas such as CV and NLP, but FederatedScope also provides support for graph learning researchers with a rich collection of datasets, the latest federated graph algorithms and benchmarks.
In this tutorial, you will learn:
- How to start graph learning with FederatedScope [click]
- How to reproduce the main experimental results in EasyFGL paper [click]
- How to use build-in or create a new federated graph dataset [click]
- How to run with built-in or new models [click]
- How to develop new federated graph algorithms [click]
- How to enable FedOptimizer, PersonalizedFL and FedHPO [click]
- Benchmarkcketing Federated GNN [click]
Quick start
Let’s start with a two-layer GCN on (fed) Cora to familiarize you with FederatedScope.
Start with built-in functions
You can easily run through a yaml file:
# Whether to use GPU
use_gpu: True
# Deciding which GPU to use
device: 0
# Federate learning related options
federate:
# `standalone` or `distributed`
mode: standalone
# Evaluate in Server or Client test set
make_global_eval: True
# Number of dataset being split
client_num: 5
# Number of communication round
total_round_num: 400
# Dataset related options
data:
# Root directory where the data stored
root: data/
# Dataset name
type: cora
# Use Louvain algorithm to split `Cora`
splitter: 'louvain'
dataloader:
# Type of sampler
type: pyg
# Use fullbatch training, batch_size should be `1`
batch_size: 1
# Model related options
model:
# Model type
type: gcn
# Hidden dim
hidden: 64
# Dropout rate
dropout: 0.5
# Number of Class of `Cora`
out_channels: 7
# Criterion related options
criterion:
# Criterion type
type: CrossEntropyLoss
# Trainer related options
trainer:
# Trainer type
type: nodefullbatch_trainer
# Train related options
train:
# Number of local update steps
local_update_steps: 4
# Optimizer related options
optimizer:
# Learning rate
lr: 0.25
# Weight decay
weight_decay: 0.0005
# Optimizer type
type: SGD
# Evaluation related options
eval:
# Frequency of evaluation
freq: 1
# Evaluation metrics, accuracy and number of correct items
metrics: ['acc', 'correct']
If the yaml file is named as example.yaml, just run:
python federatedscope/main.py --cfg example.yaml
Then, the FedAVG performance is around 0.87.
Start with customized functions
FederatedScope also provides register function to set up the FL procedure. Here we only provide an example about two-layer GCN on (fed) Cora, please refer to Start with your own case for details.
- Load Cora dataset and split into 5 subgraph
# federatedscope/contrib/data/my_cora.py
import copy
import numpy as np
from torch_geometric.datasets import Planetoid
from federatedscope.core.splitters.graph import LouvainSplitter
from federatedscope.register import register_data
from federatedscope.core.data import DummyDataTranslator
def my_cora(config=None):
path = config.data.root
num_split = [232, 542, np.iinfo(np.int64).max]
dataset = Planetoid(path,
'cora',
split='random',
num_train_per_class=num_split[0],
num_val=num_split[1],
num_test=num_split[2])
global_data = copy.deepcopy(dataset)[0]
dataset = LouvainSplitter(config.federate.client_num)(dataset[0])
data_local_dict = dict()
for client_idx in range(len(dataset)):
local_data = dataset[client_idx]
data_local_dict[client_idx + 1] = {
'data': local_data,
'train': [local_data],
'val': [local_data],
'test': [local_data]
}
data_local_dict[0] = {
'data': global_data,
'train': [global_data],
'val': [global_data],
'test': [global_data]
}
translator = DummyDataTranslator(config)
return translator(data_local_dict), config
def call_my_data(config, client_cfgs):
if config.data.type == "mycora":
data, modified_config = my_cora(config)
return data, modified_config
register_data("mycora", call_my_data)
- Build a two-layer GCN
# federatedscope/contrib/model/my_gcn.py
import torch
import torch.nn.functional as F
from torch.nn import ModuleList
from torch_geometric.data import Data
from torch_geometric.nn import GCNConv
from federatedscope.register import register_model
class MyGCN(torch.nn.Module):
def __init__(self,
in_channels,
out_channels,
hidden=64,
max_depth=2,
dropout=.0):
super(MyGCN, self).__init__()
self.convs = ModuleList()
for i in range(max_depth):
if i == 0:
self.convs.append(GCNConv(in_channels, hidden))
elif (i + 1) == max_depth:
self.convs.append(GCNConv(hidden, out_channels))
else:
self.convs.append(GCNConv(hidden, hidden))
self.dropout = dropout
def forward(self, data):
if isinstance(data, Data):
x, edge_index = data.x, data.edge_index
elif isinstance(data, tuple):
x, edge_index = data
else:
raise TypeError('Unsupported data type!')
for i, conv in enumerate(self.convs):
x = conv(x, edge_index)
if (i + 1) == len(self.convs):
break
x = F.relu(F.dropout(x, p=self.dropout, training=self.training))
return x
def gcnbuilder(model_config, input_shape):
x_shape, num_label, num_edge_features = input_shape
model = MyGCN(x_shape[-1],
model_config.out_channels,
hidden=model_config.hidden,
max_depth=model_config.layer,
dropout=model_config.dropout)
return model
def call_my_net(model_config, local_data):
# Please name your gnn model with prefix 'gnn_'
if model_config.type == "gnn_mygcn":
model = gcnbuilder(model_config, local_data)
return model
register_model("gnn_mygcn", call_my_net)
- Run with following command to start:
python federatedscope/main.py --cfg example.yaml data.type mycora model.type mygcn
Reproduce the main experimental results
We also provide configuration files to help you easily reproduce the results in our EasyFGL paper. All the yaml files are in federatedscope/gfl/baseline.
- Train two-layer GCN with Node-level task dataset Cora
python federatedscope/main.py --cfg federatedscope/gfl/baseline/fedavg_gnn_node_fullbatch_citation.yaml
Then, the FedAVG performance is around 0.87.
- Train two-layer GCN with Link-level task dataset WN18
python federatedscope/main.py --cfg federatedscope/gfl/baseline/fedavg_gcn_minibatch_on_kg.yaml
Then, the FedAVG performance is around hits@1: 0.30, hits@5: 0.79, hits@10: 0.96.
- Train two-layer GCN with Graph-level task dataset HIV
python federatedscope/main.py --cfg federatedscope/gfl/baseline/fedavg_gcn_minibatch_on_hiv.yaml
Then, the FedAVG performance is around accuracy: 0.96 and roc_aucs: 0.62.
DataZoo
FederatedScope provides a rich collection of datasets for graph learning researchers, including real federation datasets as well as simulated federation datasets split by some sampling or clustering algorithms. The dataset statistics are shown in the table and more datasets are coming soon:
| Task | Domain | Dataset | Splitter | # Graph | Avg. # Nodes | Avg. # Edges | # Class | Evaluation |
|---|---|---|---|---|---|---|---|---|
| Node-level | Citation network | Cora [1] | random&community | 1 | 2,708 | 5,429 | 7 | ACC |
| Citation network | CiteSeer [2] | random&community | 1 | 4,230 | 5,358 | 6 | ACC | |
| Citation network | PubMed [3] | random&community | 1 | 19,717 | 44,338 | 3 | ACC | |
| Citation network | FedDBLP [4] | meta | 1 | 52,202 | 271,054 | 4 | ACC | |
| Link-level | Recommendation System | Ciao [5] | meta | 28 | 5,875.68 | 20,189.29 | 6 | ACC |
| Recommendation System | Taobao | meta | 3 | 443,365 | 2,015,558 | 2 | ACC | |
| Knowledge Graph | WN18 [6] | label_space | 1 | 40,943 | 151,442 | 18 | Hits@n | |
| Knowledge Graph | FB15k-237 [6] | label_space | 1 | 14,541 | 310,116 | 237 | Hits@n | |
| Graph-level | Molecule | HIV [7] | instance_space | 41,127 | 25.51 | 54.93 | 2 | ROC-AUC |
| Proteins | Proteins [8] | instance_space | 1,113 | 39.05 | 145.63 | 2 | ACC | |
| Social network | IMDB [8] | label_space | 1,000 | 19.77 | 193.06 | 2 | ACC | |
| Multi-task | Mol [8] | multi_task | 18,661 | 55.62 | 1,466.83 | - | ACC |
Dataset format
Let’s start Dataset with torch_geometric.data. Our DataZoo contains three levels of tasks which are node-level, link-level and graph-level. Different levels of data have different attributes:
- Node-level dataset
Node-level dataset contains onetorch_geometric.datawith attributes:xrepresents the node attribute,yrepresents the node label,edge_indexrepresents the edges of the graph,edge_attrrepresents the edge attribute which is optional, andtrain_mask,val_mask,test_maskare the node mask of each splits.
# Cora
Data(x=[2708, 1433], edge_index=[2, 10556], y=[2708], train_mask=[2708], val_mask=[2708], test_mask=[2708])
- Link-level dataset
Link-level dataset contains onetorch_geometric.datawith attributes:xrepresents the node attribute,edge_indexrepresents the edges of the graph,edge_typerepresents the link label,edge_attrrepresents the edge attribute which is optional,train_edge_mask,valid_edge_mask,test_edge_maskare the link mask of each splits, andinput_edge_indexis optional if the input isedge_index.T[train_edge_mask].T.
# WN18
Data(x=[40943, 1], edge_index=[2, 151442], edge_type=[151442], num_nodes=40943, train_edge_mask=[151442], valid_edge_mask=[151442], test_edge_mask=[151442], input_edge_index=[2, 282884])
- Graph-level dataset
Graph-level dataset contains severaltorch_geometric.data, and the task is to predict the label of each graph.
# HIV[0]
Data(x=[19, 9], edge_index=[2, 40], edge_attr=[40, 3], y=[1, 1], smiles='CCC1=[O+][Cu-3]2([O+]=C(CC)C1)[O+]=C(CC)CC(CC)=[O+]2')
...
# HIV[41126]
Data(x=[37, 9], edge_index=[2, 80], edge_attr=[80, 3], y=[1, 1], smiles='CCCCCC=C(c1cc(Cl)c(OC)c(-c2nc(C)no2)c1)c1cc(Cl)c(OC)c(-c2nc(C)no2)c1')
Dataloader
For node-level and link-level tasks, we use full-batch training by default. However, some large graphs can not be adopted to full-batch training due to the video memory limitation. Fortunately, we also provide some graph sampling algorithms, like GraphSAGE and GraphSAINT which are subclasses of torch_geometric.loader.
- In node-level task, you should set:
cfg.data.loader = 'graphsaint-rw' # or `neighbor`
cfg.model.type = 'sage'
cfg.trainer.type = 'nodeminibatch_trainer'
- In link-level task, you should set:
cfg.data.loader = 'graphsaint-rw'
cfg.model.type = 'sage'
cfg.trainer.type = 'linkminibatch_trainer'
Splitter
Existing graph datasets are a valuable source to meet the need for more FL datasets. Under the federated learning setting, the dataset is decentralized. To simulate federated graph datasets by existing standalone ones, our DataZoo integrates a rich collection of federatedscope.gfl.dataset.splitter. Except for meta_splitter which comes from the meta information of datasets, we have the following splitters:
- Node-level task
community_splitter: Split by clustercfg.data.splitter = 'louvain'
Community detection algorithms such as Louvain are at first applied to partition a graph into several clusters. Then these clusters are assigned to the clients, optionally with the objective of balancing the number of nodes in each client.random_splitter: Split by randomcfg.data.splitter = 'random'
The node set of the original graph is randomly split into 𝑁 subsets with or without intersections. Then, the subgraph of each client is deduced from the nodes assigned to that client. Optionally, a specified fraction of edges is randomly selected to be removed.
- Link-level task
label_space_splitter: Split by latent dirichlet allocationcfg.data.splitter = 'rel_type'
It is designed to provide label distribution skew via latent dirichlet allocation (LDA).
- Graph-level task
instance_space_splitter: Split by indexcfg.data.splitter = 'scaffold' or 'rand_chunk'
It is responsible for creating feature distribution skew (i.e., covariate shift). To realize this, we sort the graphs based on their values of a certain aspect.multi_task_splitter: Split by datasetcfg.data.splitter = 'louvain'
Different clients have different tasks.
ModelZoo
GNN
We implemented GCN [9], GraphSAGE [10], GAT [11], GIN [12], and GPR-GNN [13] on different levels of tasks in federatedscope.gfl.model, respectively. In order to run your FL procedure with these models, set cfg.model.task to node, link or graph, and all models can be instantiated automatically based on the data provided. More GNN models are coming soon!
Trainer
We provide several Trainers for different models and for different tasks.
NodeFullBatchTrainer- For node-level tasks.
- For full batch training.
NodeMiniBatchTrainer- For node-level tasks.
- For GraphSAGE, GraphSAINT and other graph sampling methods.
LinkFullBatchTrainer- For link-level tasks.
- For full batch training.
LinkMiniBatchTrainer- For link-level tasks.
- For GraphSAGE, GraphSAINT and other graph sampling methods.
GraphMiniBatchTrainer- For graph-level tasks.
Develop federated GNN algorithms
FederatedScope provides comprehensive support to help you develop federated GNN algorithms. Here we will go through FedSage+ [14] and GCFL+ [15] as examples.
- FedSage+, Subgraph Federated Learning with Missing Neighbor Generation, in NeurIPS 2021
FedSage+ try to “restore” the missing graph structure by jointly training aMissing Neighbor Generator, each client sendsMissing Neighbor Generatorto other clients, and the other clients optimize it with their own local data and send the model gradient back in order to achieve joint training without privacy leakage.
We implemented FedSage+ infederatedscope/gfl/fedsagepluswithFedSagePlusServerandFedSagePlusClient. In FederatedScope, we need to define new message types and the corresponding handler functions.
# FedSagePlusServer
self.register_handlers('clf_para', self.callback_funcs_model_para)
self.register_handlers('gen_para', self.callback_funcs_model_para)
self.register_handlers('gradient', self.callback_funcs_gradient)
Because FedSage+ has multiple stages, please carefully deal with the msg_buffer in check_and_move_on() in different states.
- GCFL+, Federated Graph Classification over Non-IID Graphs, NeurIPS 2021
GCFL+ clusters clients according to the sequence of the gradients of each local model, and those with a similar sequence of the gradients share the same model parameters.
We implemented GCFL+ infederatedscope/gfl/gcflpluswithFedSagePlusServerandFedSagePlusClient. Since no more messages are involved, we can implement GCFL+ by simply defining how to clustering clients and adding gradients to messagemodel_para.
Enable build-in Federated Algorithms
FederatedScope provides many built-in FedOptimize, PersonalizedFL and FedHPO algorithms. You can adapt them to graph learning by simply turning on the switch.
For more details, see:
- FedOptimize
- PersonalizedFL
- FedHPO
Benchmarks
We’ve conducted extensive experiments to build the benchmarks of FedGraph, which simultaneously gains
many valuable insights for the community.
Node-level task
-
Results on representative node classification datasets with
random_splitterMean accuracy (%) ± standard deviation.Cora CiteSeer PubMed Local FedAVG FedOpt FedPeox Global Local FedAVG FedOpt FedPeox Global Local FedAVG FedOpt FedPeox Global GCN 80.95±1.49 86.63±1.35 86.11±1.29 86.60±1.59 86.89±1.82 74.29±1.35 76.48±1.52 77.43±0.90 77.29±1.20 77.42±1.15 85.25±0.73 85.29±0.95 84.39±1.53 85.21±1.17 85.38±0.33 GraphSAGE 75.12±1.54 85.42±1.80 84.73±1.58 84.83±1.66 86.86±2.15 73.30±1.30 76.86±1.38 75.99±1.96 78.05±0.81 77.48±1.27 84.58±0.41 86.45±0.43 85.67±0.45 86.51±0.37 86.23±0.58 GAT 78.86±2.25 85.35±2.29 84.40±2.70 84.50±2.74 85.78±2.43 73.85±1.00 76.37±1.11 76.96±1.75 77.15±1.54 76.91±1.02 83.81±0.69 84.66±0.74 83.78±1.11 83.79±0.87 84.89±0.34 GPR-GNN 84.90±1.13 89.00±0.66 87.62±1.20 88.44±0.75 88.54±1.58 74.81±1.43 79.67±1.41 77.99±1.25 79.35±1.11 79.67±1.42 86.85±0.39 85.88±1.24 84.57±0.68 86.92±1.25 85.15±0.76 FedSage+ - 85.07±1.23 - - - - 78.04±0.91 - - - - 88.19±0.32 - - - -
Results on representative node classification datasets with
community_splitter: Mean accuracy (%) ± standard deviation.Cora CiteSeer PubMed Local FedAVG FedOpt FedProx Global Local FedAVG FedOpt FedProx Global Local FedAVG FedOpt FedProx Global GCN 65.08±2.39 87.32±1.49 87.29±1.65 87±16±1.51 86.89±1.82 67.53±1.87 77.56±1.45 77.80±0.99 77.62±1.42 77.42±1.15 77.01±3.37 85.24±0.69 84.11±0.87 85.14, 0.88 85.38±0.33 GraphSAGE 61.29±3.05 87.19±1.28 87.13±1.47 87.09±1.46 86.86±2.15 66.17±1.50 77.80±1.03 78.54±1.05 77.70±1.09 77.48±1.27 78.35±2.15 86.87±0.53 85.72±0.58 86.65±0.60 86.23±0.58 GAT 61.53±2.81 86.08±2.52 85.65±2.36 85.68±2.68 85.78±2.43 66.17±1.31 77.21±0.97 77.34±1.33 77.26±1.02 76.91±1.02 75.97±3.32 84.38±0.82 83.34±0.87 84.34±0.63 84.89±0.34 GPR-GNN 69.32±2.07 88.93±1.64 88.37±2.12 88.80±1.29 88.54±1.58 71.30±1.65 80.27±1.28 78.32±1.45 79.73±1.52 79.67±1.42 78.52±3.61 85.06±0.82 84.30±1.57 86.77±1.16 85.15±0.76 FedSage+ - 87.68±1.55 - - - - 77.98±1.23 - - - - 87.94, 0.27 - - -
Link-level task
-
Results on representative link prediction datasets with
label_space_splitter: Hits@$n$.WN18 FB15k-237 Local FedAVG FedOpt FedProx Global Local FedAVG FedOpt FedProx Global 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 1 5 10 GCN 20.70 55.34 73.85 30.00 79.72 96.67 22.13 78.96 94.07 27.32 83.01 96.38 29.67 86.73 97.05 6.07 20.29 30.35 9.86 34.27 48.02 4.12 18.07 31.79 4.66 28.74 41.67 7.80 32.46 44.64 GraphSAGE 21.06 54.12 79.88 23.14 78.85 93.70 22.82 79.86 93.12 23.14 78.52 93.67 24.24 79.86 93.84 3.95 14.64 24.47 7.13 23.38 36.60 2.20 19.21 27.64 5.85 24.05 36.33 6.19 23.57 35.98 GAT 20.89 49.42 72.48 23.14 77.62 93.49 23.14 74.64 93.52 23.53 78.40 93.00 24.24 80.18 93.76 3.44 15.02 25.14 6.06 25.76 39.04 2.71 18.89 32.76 6.19 25.09 38.00 6.94 24.43 37.87 GPR-GNN 22.86 60.45 80.73 26.67 82.35 96.18 24.46 73.33 87.18 27.62 81.87 95.68 29.19 82.34 96.24 4.45 13.26 21.24 9.62 32.76 45.97 2.01 9.81 16.65 3.72 15.62 27.79 10.62 33.87 47.45
Graph-level task
-
Results on representative graph classification datasets: Mean accuracy (%) ± standard deviation.
PROTEINS IMDB Multi-task Local FedAVG FedOpt FedProx Global Local FedAVG FedOpt FedProx Global Local FedAVG FedOpt FedProx Global GCN 71.10±4.65 73.54±4.48 71.24±4.17 73.36±4.49 71.77±3.62 50.76±1.14 53.24±6.04 50.49±8.32 48.72±6.73 53.24±6.04 66.37±1.78 65.99±1.18 69.10±1.58 68.59±1.99 - GIN 69.06±3.47 73.74±5.71 60.14±1.22 73.18±5.66 72.47±5.53 55.82±7.56 64.79±10.55 51.87±6.82 70.65±8.35 72.61±2.44 75.05±1.81 63.40±2.22 63.33±1.18 63.01±0.44 - GAT 70.75±3.33 71.95±4.45 71.07±3.45 72.13±4.68 72.48±4.32 53.12±5.81 53.24±6.04 47.94±6.53 53.82±5.69 53.24±6.04 67.72±3.48 66.75±2.97 69.58±1.21 69.65±1.14 - GCFL+ - 73.00±5.72 - 74.24±3.96 - - 69.47±8.71 - 68.90±6.30 - - 65.14±1.23 - 65.69±1.55 - -
Results with PersonalizedFL on representative graph classification datasets: Mean accuracy (%) ± standard deviation. (GIN)
Multi-task FedBN [16] 72.90±1.33 ditto [17] 63.35±0.69
References
[1] McCallum, Andrew Kachites, et al. “Automating the construction of internet portals with machine learning.” Information Retrieval 2000
[2] Giles, C. Lee, Kurt D. Bollacker, and Steve Lawrence. “CiteSeer: An automatic citation indexing system.” Proceedings of the third ACM conference on Digital libraries. 1998.
[3] Sen, Prithviraj, et al. “Collective classification in network data.” AI magazine 2008.
[4] Tang, Jie, et al. “Arnetminer: extraction and mining of academic social networks.” SIGKDD 2008.
[5] Tang, Jiliang, Huiji Gao, and Huan Liu. “mTrust: Discerning multi-faceted trust in a connected world.” WSDM 2012.
[6] Bordes, Antoine, et al. “Translating embeddings for modeling multi-relational data.” NeurIPS 2013.
[7] Wu, Zhenqin, et al. “MoleculeNet: a benchmark for molecular machine learning.” Chemical science 2018.
[8] Ivanov, Sergei, Sergei Sviridov, and Evgeny Burnaev. “Understanding isomorphism bias in graph data sets.” arXiv 2019.
[9] Kipf, Thomas N., and Max Welling. “Semi-supervised classification with graph convolutional networks.” arXiv 2016.
[10] Veličković, Petar, et al. “Graph attention networks.” ICLR 2018.
[11] Hamilton, Will, Zhitao Ying, and Jure Leskovec. “Inductive representation learning on large graphs.” NeurIPS 2017.
[12] Xu, Keyulu, et al. “How powerful are graph neural networks?.” ICLR 2019.
[13] Chien, Eli, et al. “Adaptive universal generalized pagerank graph neural network.” ICLR 2021.
[14] Zhang, Ke, et al. “Subgraph federated learning with missing neighbor generation.” NeurIPS 2021.
[15] Xie, Han, et al. “Federated graph classification over non-iid graphs.” NeurIPS 2021.
[16] Li, Xiaoxiao, et al. “Fedbn: Federated learning on non-iid features via local batch normalization.” ICLR 2021.
[17]Li, Tian, et al. “Ditto: Fair and robust federated learning through personalization.” PMLR 2021.